Biostatistics
Generic framework for high-dimensional fixed-effects ANOVA
In functional genomics it is more rule than exception that experimental designs are used to generate the data.The samples of the resulting data sets are thus organized according to this design and for each sample many biochemical compounds are measured, e.g. typically thousands of gene-expressions or hundreds of metabolites.
Detecting regulatory mechanisms in endocrine time series measurements
The regulatory mechanisms underlying pulsatile secretion are complex, especially as it is partly controlled by other hormones and the combined action of multiple agents. Regulatory relations between hormones are not directly observable but may be deduced from time series measurements of plasma hormone concentrations. Variation in plasma hormone levels are the resultant of secretion and clearance from the circulation. A strategy is proposed to extract inhibition, activation, thresholds and circadian synchronicity from concentration data, using particular association methods.
Topology of transcriptional regulatory networks: testing and improving
With the increasing amount and complexity of data generated in biological experiments it is becoming necessary to enhance the performance and applicability of existing statistical data analysis methods. This enhancement is needed for the hidden biological information to be better resolved and better interpreted. Towards that aim, systematic incorporation of prior information in biological data analysis has been a challenging problem for systems biology.
Individual differences in metabolomics: individualised responses and between-metabolite relationships
Many metabolomics studies aim to find 'biomarkers': sets of molecules that are consistently elevated or decreased upon experimental manipulation. Biological effects, however, often manifest themselves along a continuum of individual differences between the biological replicates in the experiment. Such differences are overlooked or even diminished by methods in standard use for metabolomics, although they may contain a wealth of information on the experiment.
Inferring differences in the distribution of reaction rates across conditions
Biostatistics
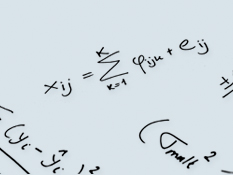
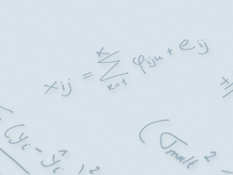
Global test for metabolic pathway differences between conditions
In many metabolomics applications there is a need to compare metabolite levels between different conditions, e.g., case versus control. There exist many statistical methods to perform such comparisons but only few of these explicitly take into account the fact that metabolites are connected in pathways or modules. Such a priori information on pathway structure can alleviate problems in, e.g., testing on individual metabolite level.
Beethoven's deafness and his three styles
Data-processing strategies for metabolomics studies
Metabolomics studies aim at a better understanding of biochemical processes by studying relations between metabolites and between metabolites and other types of information (e.g., sensory and phenotypic features). The objectives of these studies are diverse, but the types of data generated and the methods for extracting information from the data and analysing the data are similar. Besides instrumental analysis tools, various data-analysis tools are needed to extract this relevant information. The entire data-processing workflow is complex and has many steps.